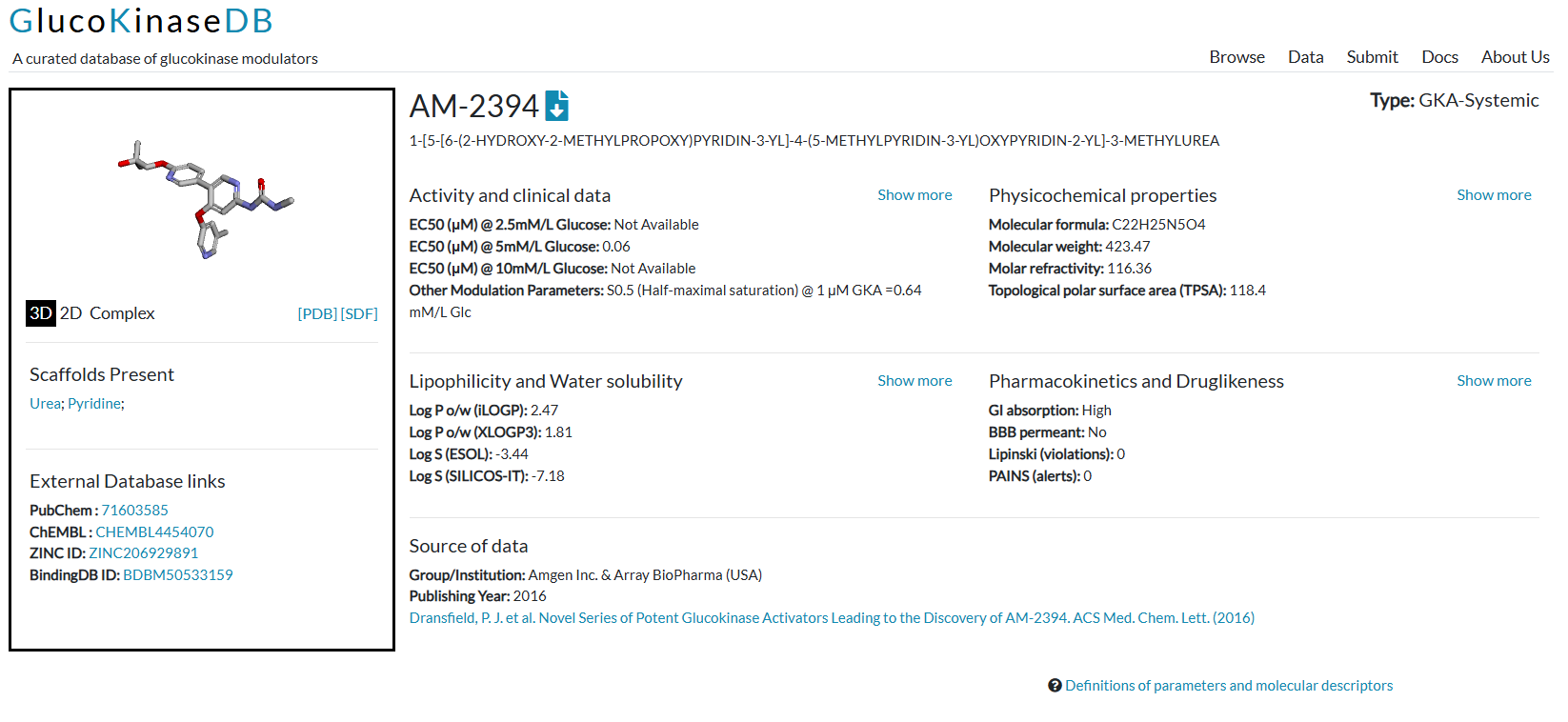
Introduction to GlucoKinaseDB
peptide-database
database
api
gkdb
glucokinase
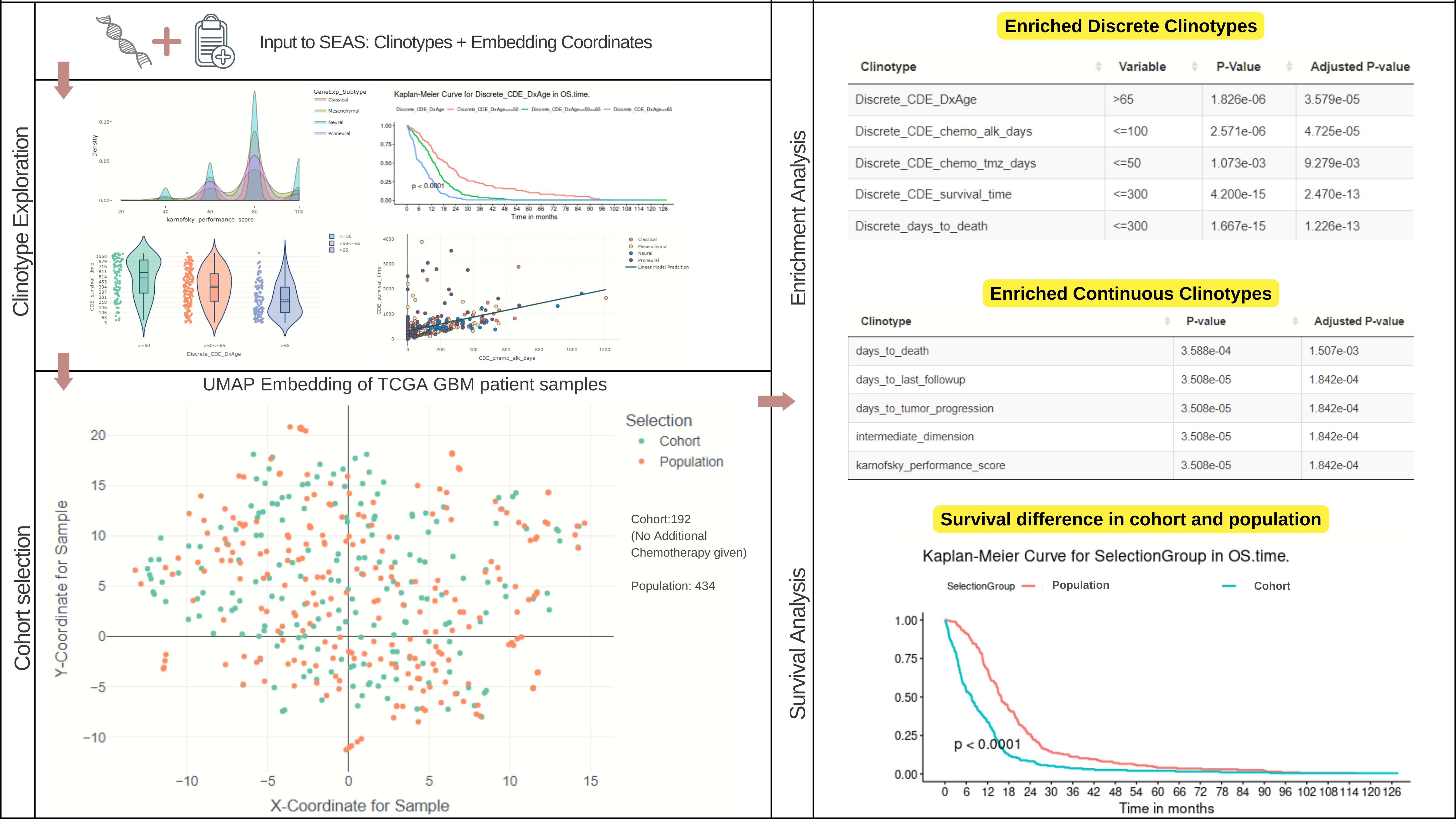
Introduction to SEAS
enrichment-analysis
seas
R
shiny
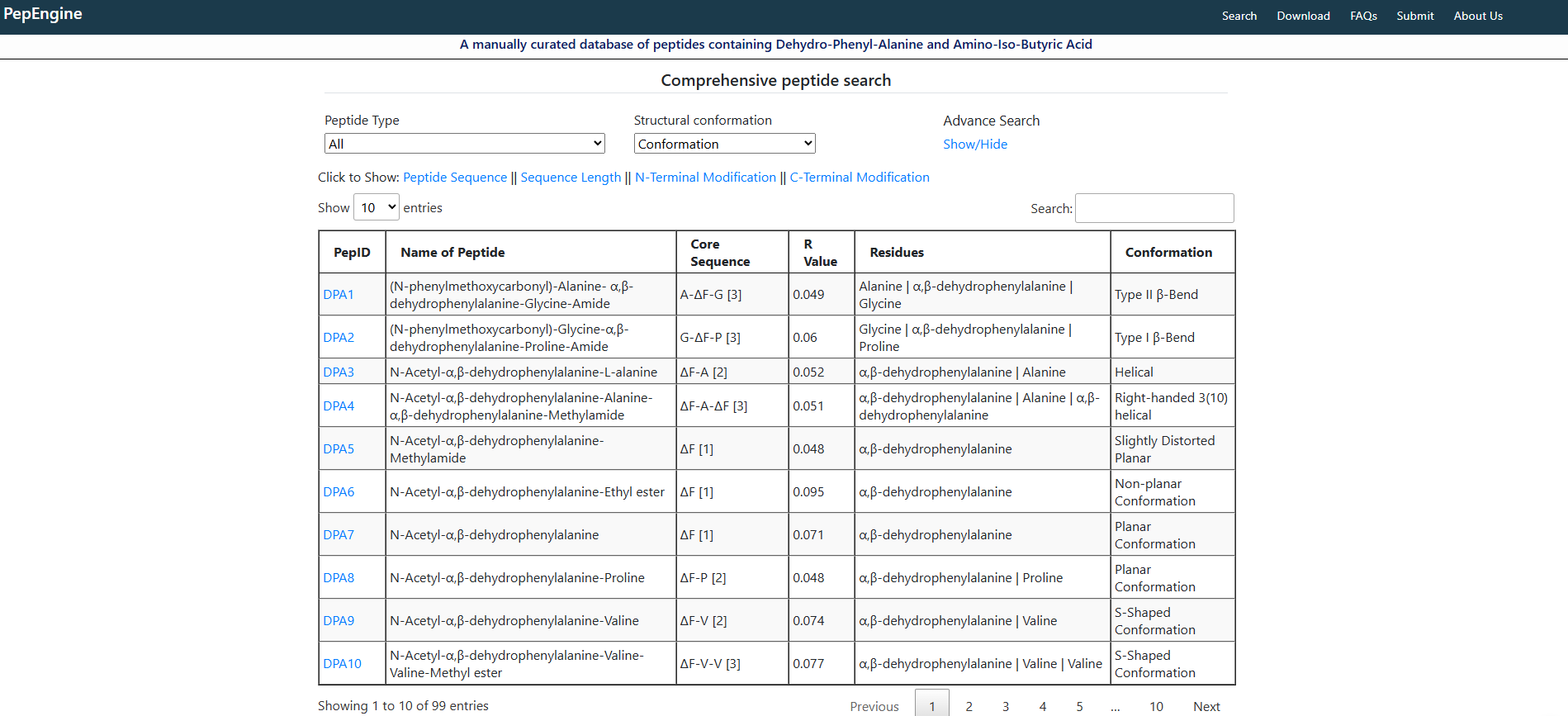
Introduction to PepEngine
peptide-database
database
pepengine
Enrichment analysis and annotation using PAGER
pathway
gene
networks
annotated-lists
cancer
signatures
cell-type
database
r
shiny
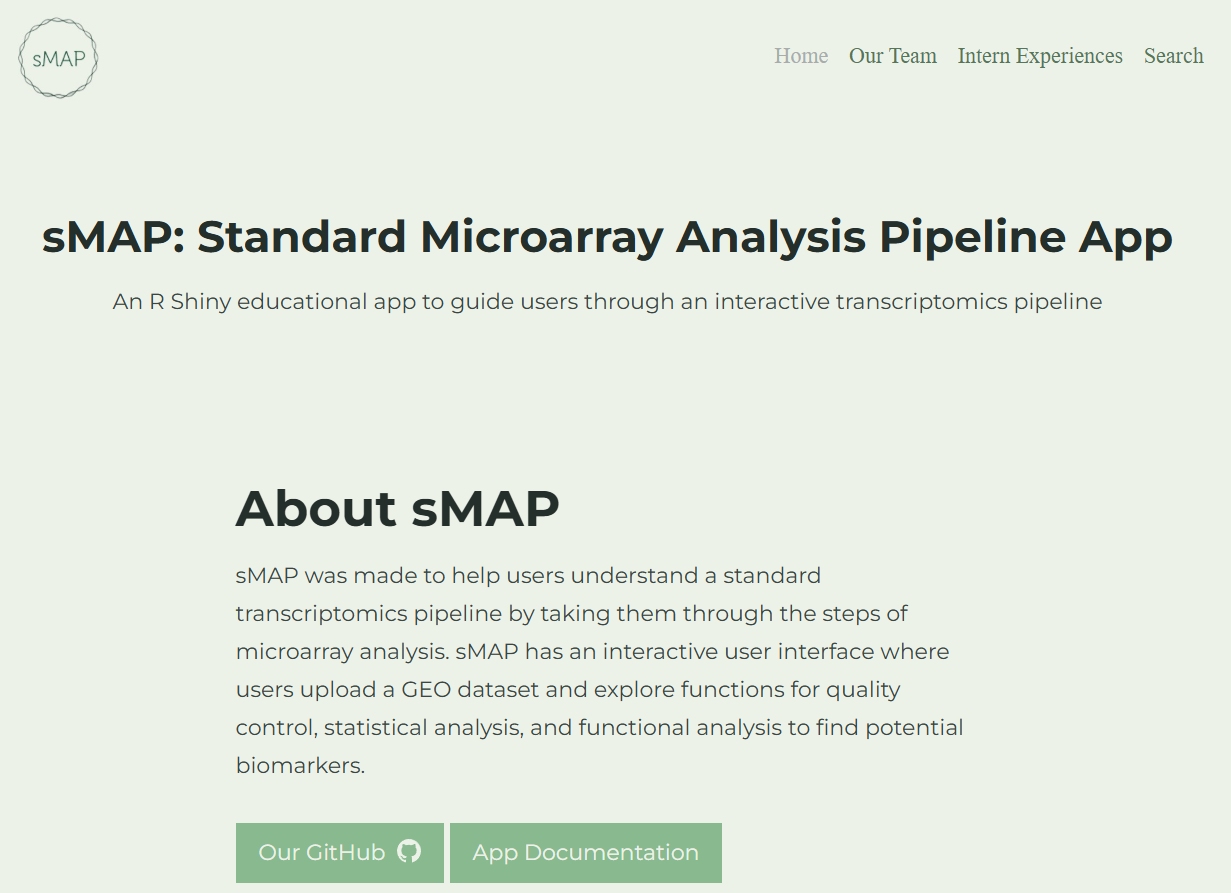
Microarray analysis for non-coders using sMAP
pathway
gene
cancer
tool
r
shiny
Deploy your Shiny App on AWS
aws
R
shiny
deployment
docker
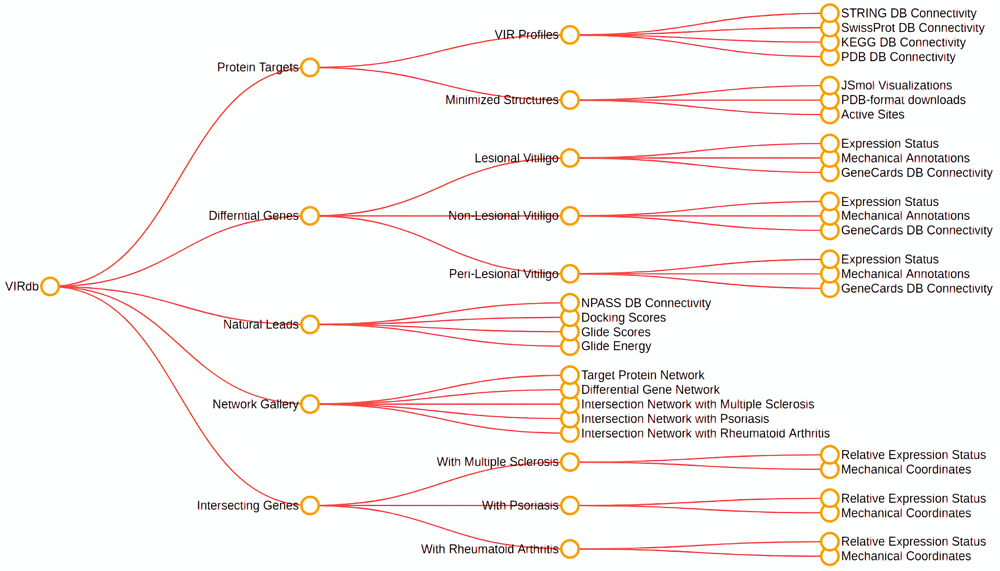
Introduction to Vitiligo Information Resource (VIRdb 2.0)
vitiligo
gene-networks
degs
protein-networks
database
No matching items